From MdsWiki
Introduction
MDSplus provided in the past an Application Programming Interface (API) for handling data in pulse files in Fortran and C. While the Fortran API is limited to reading and writing data, the C API is much more complete and allows full data and pulse file structure management (the kernel of MDSplus is based on it). However the C API is very complex, and uses a very large set of data structures, called descriptors, to handle the variety of data types available in MDSplus. For this reason, a new Object Oriented (OO) multi language API has been developed in 2008 in order to provide full data and pulse file manipulation using a much simpler interface. Currently three Object Oriented languages are supported in MDSplus: C++, Java and Python. A direct consequence of the availability of Java classes for the MDSplus object is that it is possible to use them directly from MATLAB. MATLAB in fact allows a direct mapping to imported Java classes.
A similar API in Fortran 2003 is under development, and for the moment Fortran programmers can only use the previous API, described in a following section Accessing MDSplus data in Fortran and IDL.
In this section we shall introduce the OO interface, that is, the classes and their methods defined to manipulate data in MDSplus. Regardless of the language used, the set of classes is always the same, and therefore in the general presentation we shall speak about class names, with no reference to any particular language. When code examples will be provided, the code for the three languages (C++, Java and Python) will be displayed. Language-specific issues will be then described in the last sections.
Finally, some complete examples will be presented. These examples are available in the MDSplusTutorial project presented in the first section of this tutorial, and can be run using the same pulse file provided in that project.
The Basic Classes for Accessing MDSplus Trees: Tree and TreeNode
The first class we shall see is the 'Tree' class, which describes an MDSplus tree. The constructor of Tree takes two arguments: the experiment name and the shot number. A third optional argument defines the mode of operation which can be:
- NORMAL (default mode): the tree is open for read/write. Tree editing (i.e. changing the tree structure) is not allowed;
- READONLY: the tree is open read only;
- NEW: a new tree is being created from scratch;
- EDIT: the tree is open for read/write and can also be edited, i.e. its structure can be changed.
Upon instantiation the corresponding MDSplus tree is open. It is however not possible to access data in the tree directly from the Tree class, but instead via class 'TreeNode'. TreeNode describes a single item in the MDSplus tree. TreeNode instances are returned by the Tree class via method getNode(), whose string argument is the path name of the corresponding data item. For example, with reference to the sample my_tree experiment used in the previous sections, a TreeNode instance can be derived as follows:
C++:
Tree *myTree = new Tree("my_tree", -1);
TreeNode *node1 = myTree->getNode("NUM1");
Java:
Tree myTree = new Tree("my_tree", -1);
TreeNode node1 = myTree.getNode("NUM1");
Python:
myTree = Tree('my_tree', -1)
node1 = myTree.getNode('NUM1')
Every path name whose syntax is supported by MDSplus can be passed as an argument to Tree method getNode(). Pay extra attention when using tag names: since they begin with a backslash, you must type it twice in the String argument of method getNode, e.g. myTree.getNode("\\MY_TAG").
Path names in MDSplus can be absolute or relative. In the case where you want to use relative path names, you must first define the default position in the tree, via Tree method setDefault(). This method receives as an argument a TreeNode instance, representing the new default node in the tree. For example, to get the TreeNode instance for node SUB1:SUB_NODE1, instead of using the full path name, we may do as follows:
C++:
TreeNode *sub = myTree->getNode("SUB1");
myTree->setDefault(sub);
TreeNode *subNode = myTree->getNode("SUB_NODE1");
Java:
myTree.setDefaults(myTree.getNode("SUB1"));
TreeNode subNode = myTree.getNode("SUB_NODE1")
Python:
myTree.setDefault(myTree.getNode('SUB1'))
subNode = myTree.getNode('SUB_NODE1')
The Data Framework
Let's consider the following code snippet, in which the content of tree node NODE1 is copied in node NODE2 within the experiment model my_tree.
C++
Tree *myTree = new Tree("my_tree", -1);
TreeNode *node1 = myTree->getNode("NODE1");
TreeNode *node2 = myTree->getNode("NODE2");
Data *currVal = node1->getData();
node2->putData(currVal);
Java
Tree myTree = new Tree("my_tree", -1);
TreeNode node1 = myTree.getNode("NODE1");
TreeNode node2 = myTree.getNode("NODE2");
Data currVal = node1.getData();
node2.putData(currVal);
Python
myTree = Tree('my_tree', -1)
n1 = myTree.getNode('NODE1')
n2 = myTree.getNode('NODE2')
d = n1.getData()
n2.putData(d)
The code is self-explanatory: the content of NODE1 is read into a Data instance and then written into NODE2. You may however wonder what is the actual type of data transferred: it is not specified anywhere in the example code. You will also recall that in MDSplus a variety of data types is supported, and a data item may also be a very complicated piece of element, such as the result of the compilation of a complex TDI expression. So, how is this managed?
The solution lies in this tautology: every data item is a piece of Data, which gets translated in the Object Oriented terminology into: every data class is a subclass of the generic Data class. Therefore, TreeNode method getData() will return in general a data instance.
Getting Native Types from MDSplus Data Objects
Even if a data item may be represented in MDSplus by a possible complex type, this knowledge is not required in normal operation. Normally, when reading data stored in a tree, we are interested in getting the final result, regardless of the way data have been stored. So, for example, if we want to retrieve a data item which is expected to represent an array of float values, we would like to do it directly. This is carried out by the MDSplus object framework by defining a set of basic data conversion methods in the Data class itself. They are the following:
- getByte() Convert the Data instance into a byte integer
- getShort() Convert the Data instance into a short integer
- getInt() Convert the Data instance into an integer
- getLong() Convert the Data instance into a long integer
- getFloat() Convert the Data instance into a single precision float
- getDouble() Convert the Data instance into a double precision float
- getString() Convert the Data instance into a string
and, when we are expecting arrays:
- getByteArray() Convert the Data instance into a byte integer array
- getShortArray() Convert the Data instance into a short integer array
- getIntArray() Convert the Data instance into an integer array
- getLongArray() Convert the Data instance into a long integer array
- getFloatArray() Convert the Data instance into a single precision float array
- getDoubleArray() Convert the Data instance into a double precision float array
Suppose that we want to read in a float array the content of tree node NODE1. This can be done as follows:
C++
Tree *myTree = new Tree("my_tree", -1);
TreeNode *node = myTree ->getNode("NODE1");
Data *nodeData = node->getData();
int numElements;
float *retArray = nodeData->getFloatArray(&numElements);
Java
Tree myTree = new Tree("my_tree", -1);
TreeNode node = myTree .getNode("NODE1");
Data nodeData = node.getData();
float [] retArray = nodeData.getFloatArray();
Observe that the above methods are useful only for typed languages, i.e. for which every variable has a type associated with it in its declaration (if we want to store data into a variable, you must convert it into the corresponding type). For Python this condition relaxed, and therefore the above type conversion methods are not strictly required.
Python
myTree = Tree("my_tree", -1)
node = myTree.getNode("NODE1")
data = node.getData()
native_data = data.data()
In the above example, the data() method returns a scalar or array of native numeric or string data.
Instantiating Concrete MDSplus Data Classes
Working with data without knowing how they are internally organized is fine as long as we want to read data and use it in our programs, but when we store data in the tree, we need to define the actual type and organization. So it is time to discover what are the Data concrete subclasses, and what do they describe. We shall not see all the MDSplus data classes here (the complete UML diagram is available here [1]), but we'll concentrate on those which are commonly used for reading and writing data in MDSplus trees.
The Data class has two main subclasses: Scalar and Array. The Scalar class is the common superclass for the classes describing a scalar value:
- Int8 byte integer
- Uint8 unsigned byte integer
- Int16 short integer
- Uint16 unsigned short integer
- Int32 integer
- Uin32 unsigned integer
- Int64 long integer
- Uint64 unsigned long integer
- Float32 single precision float
- Float64 double precision float
- String character string
The above classes define a constructor method which takes the native scalar value. Writing a scalar value, say a double precision value, in the tree is therefore implemented as follows:
C++
double dVal = 3.14;
Float32 *dData = new Float64(dVal);
Tree *myTree = new Tree("my_tree", -1);
TreeNode *node = myTree->getNode("NODE1");
node->putData(dData);
Java
Tree myTree = new Tree("my_tree", -1);
TreeNode node = myTree.getNode("NODE1");
double dVal = 3.14;
node.putData(new Float64(dVal));
Python
myTree = Tree('my_tree', -1)
n1 = myTree.getNode('NODE1')
n1.putData(Float64(3.14))
As you can see from the above examples, all that is required for creating a MDSplus Data instance starting from a native type, is to "vest" it with the corresponding MDSplus data class. TreeNode method putData() takes as argument a Data instance, which may be represented by any Data subclass.
The same philosophy holds for Array subclasses, describing arrays of native types. The Array subclasses are the following:
- Int8Array byte integer array
- Uint8Array unsigned byte integer array
- Int16Array short integer array
- Uint16Array unsigned short integer array
- Int32Array integer array
- Uin32Array unsigned integer array
- Int64Array long integer array
- Uint64Array unsigned long integer array
- Float32Array single precision float array
- Float64Array double precision float array
- StringArray character string array
The following examples will write an integer array in the tree:
C++
int *iArr = new int[4];
for(int i = 0; i < 4; i++) iArr[i] = i;
Int32Array *iArrData = new Int32Array(iArr, 4);
Tree *myTree = new Tree("my_tree", -1);
TreeNode *node = myTree->getNode("NODE1");
node->putData(iArrData);
Java
int[]iArr = new int[]{1,2,3,4};
Tree myTree = new Tree("my_tree", -1);
TreeNode node = tree.getNode("NODE1");
node.putData(new Int32Array(iArr));
Python
myTree = Tree('my_tree', -1)
n1 = myTree.getNode('NODE1')
n1.putData(Int32Array([1, 2, 3, 4]))
When writing native data types, it is even not necessary to vest the datum with the corresponding class, because method putData() is overloaded so that the passed data type is internally transformed into a MDSplus data class. So, the last line in the above examples could have been:
C++
node->putData(iArr, 4);
Java
node.putData(iArr);
Python
n1.record = [1, 2, 3, 4]
In Python, the method putData() is called implicitly when assigning a value to the record property.
References to Other Nodes in the Tree
So far we have discovered how to use scalars and arrays. We know however that data in MDSplus can be much more complex. Data can be, for example, a reference to a node in the tree, so we would expect a new Data type representing a reference to another node in the tree. We have already met this class: TreeNode. TreeNode in fact is a subclass of Data and, as such, can be used as any other data item. So, if we want to store in NODE1 a reference to NODE2 in tree my_tree, we do as follows:
C++
Tree *myTree = new Tree("my_tree", -1);
TreeNode *node1 = myTree->getNode("NODE1");
TreeNode *node2 = myTree->getNode("NODE2");
node1->putData(node2);
Java
Tree myTree = new Tree("my_tree", -1);
TreeNode node1 = myTree.getNode("NODE1");
TreeNode node2 = myTree.getNode("NODE2");
node1.putData(node2);
Python
myTree = Tree('my_tree', -1)
node1 = myTree.getNode('NODE1')
node2 = myTree.getNode('NODE2')
node1.putData(node2)
In the above examples, we have used an already resolved node reference. It may happen, however, that a reference to a node not yet resolved (e.g. belonging to another tree which is not linked yet). In this case, we can use class TreePath, whose constructor takes a string argument, i.e. the pathname, or tagname, of the referenced node.
How to Build More Complex Expressions
In the MDSplus internals, a generic expression is represented by a tree of data instance, in a way which is similar to the construction of syntax trees when parsing a structured expression in compilers. For example, the expression 2 + NODE1 is internally represented by a tree whose root describes the add operation and which has two children: the first one describes the integer and the second one the reference to NODE1. This internal organization is reflected in the corresponding hierarchy of class instances, and the expression will be represented by a object tree where the root of class Function describes the operation and its two children are represented by an instance of Int32 and TreeNode, respectively. The representation of this expression may be built and written in node NODE2 of my_tree as follows:
C++
Tree *myTree = new Tree("my_tree", -1);
TreeNode *node1 = myTree->getNode("NODE1");
TreeNode *node2 = myTree->getNode("NODE2");
Data *args[2];
args[0] = new Int32(2);
args[1] = node1;
Function *expr = new Function(38, 2, args);
node2->putData(expr);
Java
Tree myTree = new Tree("my_tree", -1);
TreeNode node1 = myTree.getNode("NODE1");
TreeNode node2 = myTree.getNode("NODE2");
Function expr = new Function(38, new Data[]{new Int32(2), node1});
//The constructor of class Function takes the opcode (38 means addition)
//and an array of children nodes.
node2.putData(expr); //The expression is written in my_tree
Python
myTree = Tree("my_tree", -1)
node1 = myTree.getnode("NODE1")
node2 = myTree.getNode("NODE2")
node2.putData(Function("add", (2, node1)))
Not a very user friendly way of building expressions, isn't it?
There is however no need to know all this for using expressions, as the Data class defines a static compile() method which takes a TDI expression as string argument and returns the corresponding Data object hierarchy describing the complete expression. The returned data may be a simple scalar instance or a big tree in memory of class instances: we do not care, as all this is transparent to us!
The following example builds the same expression of the previous example and stores it in NODE2, using the compile() method.
C++
Tree *myTree = new Tree("my_tree", -1);
TreeNode *node2 = myTree->getNode("NODE2");
Data *expr = compile("2+NODE1");
node2->putData(expr);
Java
Tree myTree = new Tree("my_tree", -1);
TreeNode node2 = myTree.getNode("NODE2");
Data expr = Data.compile("2+NODE1");
node2.putData(expr);
Python
myTree = Tree('my_tree', -1)
node2 = myTree.getNode('NODE2')
node2.putData(myTree.tdiCompile("2 + NODE1"))
Using a string to describe an expression is not always enough. Suppose, for example,that we want to store in my_tree (node NODE1) an expression describing an acquired raw short array of 1000 samples (in program variable rawArr) which gets added by an offset of 5 and multiplied by a gain of 3, and whose timebase is described by another vector of 1000 single precision float values (in program variable dimArr). This can be done by replacing the raw and dimension field in the BUILD_SIGNAL expression (refer to the first section of this tutorial), by symbols $1 and $2, and providing the variables as added arguments in method compile:
C++
short rawArr[1000];
float dimArr[1000];
//Fill rawArr and dimArr
...
Tree *myTree = new Tree("my_tree", -1);
TreeNode *node1 = myTree->getNode("NODE2");
Int16Array *rawData = new Int16Array(rawArr, 1000);
Float32Array *dimData = new Float32Array(dimArr, 1000);
Data *expr = compileWithArgs("BUILD_SIGNAL(($VALUE + 5)*3, $1, $2)", 2, rawData, dimData);
//$VALUE in signals means the raw data
//In C++ the additional arguments must be preceded by their number
node1->putData(expr);
Java
short []rawArr = new short[100];
float []dimArr = new float[1000];
//Fill rawArr and dimArr
...
Tree myTree = new Tree("my_tree", -1);
TreeNode node1 = myTree.getNode("NODE1");
Data expr = Data.compile("BUILD_SIGNAL(($VALUE + 5)*3, $1, $2)",
new Data[]{new Int16Array(rawArr), new Float32Array(dimArr)})
//In Java the additional arguments are passed as a Data array
node1.putData(expr);
Python
myTree = Tree('my_tree', -1)
node1 = myTree.getNode('NODE1')
expr = myTree.tdiCompile("BUILD_SIGNAL(($VALUE + 5) * 3, $1, $2)", rawData, dimData)
node1.putData(expr)
Printing MDSplus Data Objects
We have seen how it is possible to manage a variety of different data types representing MDSplus data in Object Oriented languages in an elegant way. Most of the times, in fact, we are not interested in the actual organization of data and the set of generic methods defined for the Data class suffice for our purposes. So, we can even forget about the actual MDSplus data classes and work with generic Data objects, letting the overriding mechanism do the real stuff. For example, when we read from the pulse file a data item describing the acquired waveform of some interesting physical phenomena, we are not concerned about the way data have been internally organized, and all we want is to have the sample array in a, say, float array variable. So we read the signal, getting a Data object (actually something more complicated, possibly a hierarchy of object instances, but we do not care), then we call method getFloatArray() for the returned data object and we are done.
When handling generic objects in our programs, it is often useful to convert them in some textual format, e.g. to print information messages for debugging. A method defined in the Data class (and therefore available in all data objects) is decompile() which returns the textual description of the actual object, represented by the corresponding TDI expression. There will be in practice little need to explicitly call the method decompile(), since this is implicitly called by the language environment when a Data object is printed. In C++ this happens when sending the object to a cout stream, in Java and Python this is done by the implicit String conversion carried out by methods toString() and __str__(), respectively. The following example will show how the textual representation of an Data object read from my_tree gets printed in the standard output.
C++
Tree *myTree = new Tree("my_tree", -1);
TreeNode *node1 = myTree->getNode("NODE2");
Data *data = node1.getData();
cout << "Data read from NODE1: " << data << "\n";
Java
Tree myTree = new Tree("my_tree", -1);
TreeNode node1 = myTree.getNode("NODE1");
Data data = node1.getData();
System.out.println("Data read from NODE1: " + data);
Python
myTree = Tree('my_tree', -1)
node1 = myTree.getNode('NODE1')
data = node1.getData()
print 'Data read from NODE1: ', data
Note that the textual description returned by decompile() may be very long in the case of large arrays, and is intentionally left incomplete when the dimension is above a given value.
Accessory Data Information
Accessory information can be associated with every Data object. Four types of accessory information are available:
- HELP for associating a description with the data item
- UNITS to describe the units used
- ERROR to describe the error associated with the data item
- VALIDATION to describe a validation for this data
Any Data object can be used to describe any accessory information, which gets stored with the data item in the tree. Usually, the help information is represented by a String object providing some sort of documentation for that data. Units, as well, are normally represented by strings describing the units used (they are recognized by jScope and displayed in the corresponding axis). Error and validation fields may be represented by other data types: errors may be represented by a list of two arrays (refer to the Lists section below) describing the limits of the error bar associated with every element of an array, and validation may be represented by an expression which returns a quality index for the associated data.
Accessory data can be accessed via Data accessor methods: setHelp(Data), getHelp(), setUnits(Data), getUnits(), setValidation(Data), getValidation(), setError(Data), getError().
When getting the decompiled version of a data item with some accessory information, you will get a somewhat complicated TDI expression. Look for example at the following snapshot of Python:
d = Int32(1)
d.setUnits('Ohm')
print d
Build_With_Units(1, "Ohm")
The accessory unit information is embedded in the data instance tree to form a new expression: Build_With_Units(1, "Ohm").
In Java you would need to convert the units string to an MDSplus data type as follows:
Java
d=Int32(1)
d.setUnits(new MDSplus.String("Ohm"))
C++
Data *d = new Int32(1);
d->setUnits(new String("Ohm"));
Even though long supported by MDSplus, accessory information has been rarely used since they led to awkward expressions. The new interface provides a much easier way of dealing with such information.
More on Arrays
We have seen so far how arrays are represented by a set of Array classes. The same classes allow the representation of multidimensional arrays. The constructor method of the Array classes in fact allow the dimensions to be specified together with the array data. The following example shows how a bi-dimensional array is created to describe the 3x2 integer array [[1,2],[11,22],[111,222]]
C++
int arr[6]={1,2,11,22,111,222};
int dims[2] = {3,2};
Data *arrD = new Int32Array(arr, 2, dims);
Java
int arr[] = new int[]{1,2,11,22,111,222};
int dims[] = new int[]{3,2};
Data arrD = new Int32Array(arr, dims);
Python
arrD = Int32Array([[1, 2], [11, 22], [111, 222]])
It is always possible to inspect the dimensionality of any data, via Data method getShape() which return a native integer array describing data dimensions. Be aware that only arrays support the concept of dimension so in general method getShape() may generate an error.
In python, one can also obtain a numpy array instance from an MDSplus array instance simply by calling the data() method of the MDSplus array instance:
arrD = Int32Array([[1, 2], [11, 22], [111, 222]]) npa = arrD.data() type(npa) <type 'numpy.ndarray'>
It is possible to get and set single elements or subarrays of any Array object via methods getElementAt() and setElementAt(), respectively. Both methods take as an argument an array of integer values which specify the (possibly incomplete) index of the element. For example, considering the 3x2 array of the above example, method getElementAt([2, 0]) will return a Int32 object containing the number 222, and method getElementAt([2]) will return a Int32Array object containing the array [111, 222].
Arrays are stored and retrieved using Row-Major ordering, that is, rows are stored in subsequent memory locations. This is the normal way multidimensional arrays are stored in C and Java, BUT NOT in Fortran that uses Column-Major ordering.
Multi dimensional arrays are consistently stored and retrieved using the Python, C++ and Java interfaces, but are interpreted in a different way when accessed via TDI. TDI uses Column-Major ordering and therefore unexpected results may occur when mixing the usage of Java, C++ and Python and TDI.
The following python example will clarify a bit, and possibly save a big headache.
Suppose a tree named TEST containing a node named M_ARRAY:
t = Tree('TEST', -1)
n = t.getNode('M_ARRAY')
array = Int32Array([[1, 2], [11, 22], [111, 222]]) #three rows of two columns each
n.putData(array) #save array
d = n.data() #read back array
d[0,1] #get first row, second column
2 #as expected
Data.execute('M_ARRAY[0,1]') # Evaluate the TDI expression specifying element (0,1) of node M_ARRAY
11 # why???? because TDI uses Column-Major ordering, so second row, first column is retrieved
Data.execute('M_ARRAY[1,0]') #This is the correct way for getting first row, second column
2 # yielding the correct result
Manipulating Trees
Common Operations
We have already seen how classes Tree and TreeNode are used to access the elements of an MDSplus tree. The set of methods available for these classes is very rich and allows all the operations which are supported by MDSplus. In the following we shall describe those methods which are more frequently used. For a complete description you may refer to the UML class representation or to the language-specific references.
The most common TreeNode methods are getData() and putData(). getData() returns the data stored in that node as a Data instance. putData takes a Data instance as argument and stores that data in the associated node of the tree.
A tree node, represented by a TreeNode instance can be queried for a variety of information, among which its pathname (methods getMinPath(), getPath(), getFullPath()) , the size of the contained data (method getLength()), the node usage (method getUsage()) its current ON/OFF state (method isOn()), the parent node (method getParent()) and the list of the tag names associated with that node (method getTags()).
It is also possible to get the list of descendants for a given tree node, via Tree method getNodeWild(), taking as argument a wildcard name and, optionally, the selected usage. This method will likely return a list of TreeNode instances, for which some common operation will be then performed (e.g. getting the path name). For this reason, the support class TreeNodeArray has been defined, which has a set of accessor methods is defined. The accessor methods have the same name of the corresponding accessor TreeNode method, with the difference that it will return/accept an array of native types, instead a single value. The usage of TreeNodeArray is clarified by the following example, which prints the path name of all the nodes of tree my_tree.
C++
Tree *my_tree = new Tree("my_tree", -1);
TreeNodeArray *nodeArr = my_tree->getNodeWild("***");
StringArray *names = nodeArr->getPath();
//method getPath returns a MDSplus StringArray type describing an array of strings
//(C++ has no native type for this)
for(int i = 0; i < nodeArray->getNumNodes(); i++)
cout << names->getElementAt(i) << "\n";
Java
Tree my_tree = new Tree("my_tree", -1);
TreeNodeArray nodeArr = my_tree.getNodeWild("***");
java.lang.String [] names = nodeArr.getPath();
//Array of Strings are natively supported by Java
for(int i = 0; i < names.length; i++)
System.out.println(names[i]);
Python
my_tree = Tree('my_tree', -1)
nodeArr = my_tree.getNodeWild('***')
print nodeArr.getPath()
Handling Multiple Trees
An important feature of MDSplus Tree and TreeNode objects is the possibility of handling multiple open trees at the same time. Every new Tree instance in fact refers to a experiment and shot passed to the constructor method. When a TreeNode instance is retrieved by a given Tree instance, that TreeNode refers to that tree. Operations on TreeNode instances can be mixed, and every TreeNode will redirect the selected operation to the tree it refers to.
Editing Trees
In order to modify the structure of a MDSplus tree (e.g adding tags, adding, renaming or removing nodes) it is necessary to open the tree in EDIT mode. If a Tree is being built from scratch, it is necessary to open it in NEW mode. The mode (String) argument is passed to the Tree constructor. The following methods are commonly used when editing trees:
- addNode(String name, String usage) add a new node to the tree (Tree method)
- deleteNode(String name) remove a node and its descendants from the tree (Tree method)
- renameNode(String oldName, String newName) rename a given tree node (Tree method)
- addTag(String tagName) add a tag name to the node (TreeNode method)
- removeTag(String tagName) remove a tag from the tag list associated with this node (TreeNode method)
- write() write the tree being edited on disk (Tree method)
The following example shows how tree my_tree used in the first section of this tutorial, adding also a tag to the last created node.
C++
Tree *tree = new Tree("my_tree", -1, "NEW");
tree->addNode(":NUM1", "NUMERIC");
tree->addNode(":NUM2", "NUMERIC");
tree->addNode(":NUM3", "NUMERIC");
tree->addNode(":TXT", "TEXT");
TreeNode *sub1Node = tree->addNode(".SUB1", "STRUCTURE");
tree->setDefault(sub1Node);
tree->addNode(":SUB_NODE1", "NUMERIC");
tree->addNode(".SUB2", "STRUCTURE");
TreeNode *lastNode = tree->addNode(".SUB2:SUB_NODE2", "NUMERIC");
lastNode->addTag("TAG1");
tree->write();
Java
Tree tree = new Tree("my_tree", -1, "NEW");
tree.addNode(":NUM1", "NUMERIC");
tree.addNode(":NUM2", "NUMERIC");
tree.addNode(":NUM3", "NUMERIC");
tree.addNode(":TXT", "TEXT");
tree.setDefault(tree.addNode(".SUB1", "STRUCTURE"));
tree.addNode(":SUB_NODE1", "NUMERIC");
tree.addNode(".SUB2", "STRUCTURE");
TreeNode lastNode = tree.addNode(".SUB2:SUB_NODE2", "NUMERIC");
lastNode.addTag("TAG1);
tree.write();
Python
tree = Tree('my_tree', -1, 'NEW')
tree.addNode(":NUM1", "NUMERIC")
tree.addNode(":NUM2", "NUMERIC")
tree.addNode(":NUM3", "NUMERIC")
tree.addNode(":TXT", "TEXT")
tree.setDefault(tree.addNode(".SUB1", "STRUCTURE"))
tree.addNode(":SUB_NODE1", "NUMERIC")
tree.addNode(".SUB2", "STRUCTURE")
tree.addNode(".SUB2:SUB_NODE2", "NUMERIC").addTag("TAG1")
tree.write()
Another Tree method which is useful in practice is method createPulse(int shot) which creates a new pulse file starting from the experiment model the Tree instance refers to.
Using MDSplus Segments for Continuous Data Acquisition
When a data item is written in a MDSplus tree, the previous content is replaced by the new one. It is however possible to use a different storage strategy, i.e. to append a new chunk of data to what is currently stored in the tree node. This policy is used, for example, to handle a long data acquisition, where new blocks of data become available from time to time. At any time the data currently stored in the tree is available for readout.
Using MDSplus Segments
Continuous data acquisition is achieved in MDSplus via the Segment concept. New segments of data can be appended to the stored segment sequence. Every segment is identified by the following items:
- start the start time for the segment
- end the end time for the segment
- dimension the dimension for the segment, i.e. the time associated with segment samples.
- data the segment data. Data has to be a N-dimensional Array, meaning that a N-1 dimensional array is associated with every time sample. In the simplest case, data is a 1-dimensional array, where a scalar value is associated with every time sample.
While the data for a given segment have to be arrays (i.e. they must be represented by a subclass of Array), generic expressions can be used for start, end and dimension. The start and end information are then used to efficiently retrieve portions of stored signals, i.e. without reading the whole data set, but only the segments belonging to the selected time range (see below on how retrieving portions of large data).
The following example shows how to append to tree node SIG_1 a new data segment describing a 1 second data acquisition from time 10 to time 11 at 1 kHz sampling speed.
C++
float data[1000];
Tree *tree = new Tree("my_tree", -1);
TreeNode *node = tree->getNode("SIG_1");
///Fill previous segments
...
//acquire new set of 1000 samples in data
...
Data *start = new Float64(10.);
Data *end = new Float64(11.);
Data *dimension = new Range(start, end, new Float64(1E-3));
//Dimension is represented by a MDSplus Range data object. Range describes a
//single speed clock by specifying the init and
//end time and the clock period.
Array *dataArray = new Float32Array(data, 1000);
node->beginSegment(start, end, dimension, data);
Java
float []data = new float[1000];
Tree tree = new Tree("my_tree", -1);
TreeNode node = tree.getNode("SIG_1");
///Fill previous segments
...
//acquire new set of 1000 samples in data
...
Data start = new Float64(10.);
Data end = new Float64(11.);
Data dimension = new RangeData(start, end, new Float64(1E-3));
//Dimension is represented by a MDSplus Range data object. Range describes a
//single speed clock by specifying the init and
//end time and the clock period.
Array dataArray = new Float32Array(data, 1000);
node.beginSegment(start, end, dimension, data);
Python
#assume that 1000 samples are contained in data
n = Tree('my_tree', -1).getNode('SIG_1')
start = Float64(10.)
end = Float64(11.)
dim = Range(start, end, Float64(1E-3))
n.beginSegment(start, end, dim, Float32Array(data))
Filling Segments
In the previous example we have built and stored a segment in a single operation. This is fine in the case we have in a single chunk all the segment data. There may be however situations in which not all the data for the current segment is available at the same time. For example, we may wish to declare an initialize a segment for 10000 samples, and then we may get data samples in chunks of 1000 samples. In this case, after defining the segment and appending to the node via beginSegment() method, me can fill its data content via method putSegment(). The arguments if method putSegment() are the (partial) data array, and a row index which specify the starting position of the passed data subarray. Normally this argument is -1, meaning that the portion of the data for this segment has to be appended to the end of the currently saved data segment. In this case the meaning of the data array passed to method beginSegment() represents the default value of the segment (in the case its has been only partially filled).
The following example continues the code of the previous example assuming that the segment is filled in 10 iterations, by appending a chunk of 100 samples at each iteration.
C++
//from previous example
float currData[100];
for(int i = 0; i < 10; i++)
{
//Fill currData array with acquired data chunk
Array *subArr = new Float32Array(currData, 100);
n->putSegment(subArr, -1);
}
Java
//from previous example
float []currData = new float[100];
for(int i = 0; i < 10; i++)
{
//Fill currData array with acquired data chunk
Array subArr = new Float32Array(currData, 100);
n.putSegment(subArr, -1);
}
Python
for i in range(0, 10): #fill currData with a chunk of 100 samples n.putSegment(Float32Array(currData), -1)
Using Timestamped Segments
We have seen that segments require the definition of accessory information such as the associated start and end time. When sampling time is described by a 64-bit integer, it is possible to use a more simplified interface provided by TreeNode putRow(). The arguments of methods putRow() are:
- The data sample, represented either by a scalar value or an array, which represents data associated with the sample
- The timestamp for that sample
Method putRow handles MDSplus segments internally: every time it is called, a new sample is appended the current segment, and when the segment gets filled, a new segment is created. The following example shows how a data acquisition composed of 1000 samples gets stored in a MDSplus tree.
C++
Tree *tree = new Tree("my_tree", -1);
TreeNode *node = tree->getNode("SIG_1");
for(int i = 0; i < 1000; i++)
{
float currVal;
//fill currVal (here a scalar value) with the acquired sample
//...
Float32 *currData = new Float32(currVal);
_int64 currTime = i; //Time is here the index itself
node->putRow(currData, &currTime);
}
Java
Tree tree = new Tree("my_tree", -1);
TreeNode node = tree.getNode("SIG_1");
for(int i = 0; i < 1000; i++)
{
float currVal;
//fill currVal (here a scalar value) with the acquired sample
//...
node.putRow(new Float32(currVal), (long)i);
}
Python
n = Tree('my_tree', -1).getNode('SIG_1')
for i in range(0, 1000):
#get sample in currVal
n.putRow(10000, Float32(currVal), Int64(i))
In the Python example, the dimension of the segment has been explicitly set, otherwise it defaults to 10000.
From the examples above, you can realize that the use of method putRow() for appending data in MDSplus is much more simple than handling segments in the program. There are however some drawbacks in using putRow():
- The value of the associated time can only be a 64-bit integer,
- Writing data is not very efficient, singe a single sample is stored on disk at each operation. Conversely, when using segments, blocks of data are written to disk in a single operation, and this may improve dramatically performance.
Using Segments to store images in pulse files.
MDSplus segments are useful to store sequences of images (frames) in MDSplus. Formerly, sequences of images have been stored in MDSplus as signals whose data part was represented by a 3 D arrays, where the last dimension represented time.
Storing frames in this way, however, produces soon huge signals which could be hardly managed, due to the large amount of memory resources needed for their access and visualization. In particular, jScope runs quickly out of memory resources when requested to show frame sequences.
A much better way for storing frame sequences is to use segments to store frames. Every segment can host one or more frames, so that individual (sets of) frames can be separately retrieved (segment readout) without the need of storing the whole sequence of frames into memory.
The latest version of jScope uses segmented data access, whenever frames are stored in this way, and is now capable to show long sequences of frames with a reduced consumption of memory resources. As an example, jScope has been used to display a sequence of 1920x1080 16 bit frames acquired at 10Hz for one hour!
In the following, a complete sample C++ program is presented, which stores a sequence of frames in a node tagged SEGMENTED in pulse file TEST, shot 1. The frames are all black, except for a white line whose angle changes along the sequence. In this example, every segment contains exactly one frame and the time associated with every segment is just the segment number. Storing a single frame per segment is a very common choice as the dimension of the frames is large enough to allow efficient data access. For example, a 640x480 16 bit frame 1s 614.4 kBytes large, a dimension worth an entire segment. Clearly, in a more realistic example, time associated with frames (segments) will represent the actual sampling time for that frame.
#include <cstring>
#include <math.h>
#include <mdsobjects.h>
#define WIDTH 640
#define HEIGHT 480
#define NUM_FRAMES 100
#define PI 3.14159
using namespace MDSplus;
int main(int argc, char *argv[])
{
try {
/*Open TEST pulse file, shot 1 */
Tree *t = new Tree("test", 1);
/* Get node \SEGMENTED */
TreeNode *node = t->getNode("\\segmented");
/* Make sure no data is contained */
node->deleteData();
/* Allocate space for a 16 bit frame */
short *currFrame = new short[WIDTH*HEIGHT];
/* Build and store the frame sequence */
for(int frameIdx = 0; frameIdx < NUM_FRAMES; frameIdx++)
{
/* get the angular coefficient of the current line */
double m = tan((2*PI*frameIdx)/NUM_FRAMES);
/* Prepare the current frame (black with a white line) */
memset(currFrame, 0, WIDTH * HEIGHT * sizeof(short));
for(int i = 0; i < WIDTH; i++)
{
int j = (int)round((i-WIDTH/2)*m);
if(j >= -HEIGHT/2 && j < HEIGHT/2)
currFrame[(j+HEIGHT/2)*WIDTH +i] = 255;
}
/* Time is the frame index */
float currTime = frameIdx;
/* Start time and end time for the current segment are the same (1 frame is contained) */
Data *startTime = new Float32(currTime);
Data *endTime = new Float32(currTime);
/* Segment dimension is a 1D array with one element (the segment has one row) */
int oneDim = 1;
/* Data dimension is a 3D array where the last dimension is 1 */
Data *dim = new Float32Array(&currTime, 1, &oneDim);
int segmentDims[3];
/* unlike MDSplus C uses row-first ordering, so the last MDSplus dimension becomes the first one in C */
segmentDims[0] = 1;
segmentDims[1] = HEIGHT;
segmentDims[2] = WIDTH;
Data *segment = new Int16Array(currFrame, 3, segmentDims);
/* Write the entire segment */
node->makeSegment(startTime, endTime, dim, (Array *)segment);
/* Free stuff */
deleteData(segment);
deleteData(startTime);
deleteData(endTime);
deleteData(dim);
}
}catch(const MdsException & exc)
{
std::cout << "Error appending segments: " << exc.what();
}
}
Even when a single frame is saved in a segment, it is necessary to declare its dimension as a 1D array (with one element in this case), as in general MDSplus expects a sequence of rows in each segment. For the same reason the frame is not declared as a 2D array, but as a 3D array where the last dimension is 1.
The following is the same example but this time written in Python:
from MDSplus import *
from numpy import array
import math
WIDTH = 640
HEIGHT = 480
NUM_FRAMES = 100
PI = 3.14159
tree = Tree("test",1,"NEW")
node = tree.addNode("SEGMENTED").addTag("SEGMENTED")
node = tree.getNode("\\SEGMENTED")
node.deleteData()
currFrame = array([0]*(WIDTH*HEIGHT))
for frameIdx in range(0, NUM_FRAMES):
m = math.tan((2*PI*frameIdx)/NUM_FRAMES)
for i in range(WIDTH):
j = int(round((i-WIDTH/2)*m))
if j >= -HEIGHT/2 and j < HEIGHT/2:
currFrame[(j+HEIGHT/2)*WIDTH+i] = 255
currTime = float(frameIdx)
startTime = Float32(currTime)
endTime = Float32(currTime)
dim = Float32Array(currTime)
segment = Int16Array(currFrame)
segment.resize([1, HEIGHT, WIDTH])
node.makeSegment(startTime, endTime, dim, segment)
tree.write()
To write the data with multiple frames per segment, with a dimension that approximates video frame rate use beginSegment, and putSegment. Here is and equivalent example structured this way:
from MDSplus import *
from numpy import array
import math
# some constants needed to make the frames
WIDTH = 640
HEIGHT = 480
SEG_SIZ=10
NUM_FRAMES = 128
PI = 3.14159
delta_t = 1/30.
trigger_time = 0.0
# create a new tree called test, shot 1
# and add a node to hold the segmented data
tree = Tree("test",1,"NEW")
node = tree.addNode("SEGMENTED").addTag("SEGMENTED")
node.deleteData()
tree.write()
# Open the tree and get the node to put the data in.
t = Tree('test', 1)
node = t.SEGMENTED
# make a buffer to hold the computed frames
# make a dummy buffer the size and shape of
# an ENTIRE SEGMENT
#
currFrame = array([0]*(WIDTH*HEIGHT))
currSeg = array([0]*(WIDTH*HEIGHT*SEG_SIZ))
currSeg = Int16Array(currSeg)
currSeg.resize(SEG_SIZ, HEIGHT, WIDTH)
# for each frame
for frameIdx in range(0, NUM_FRAMES):
# if this is the start of a new segment then
# call beginSegment
if frameIdx % SEG_SIZ == 0:
currTime = float(trigger_time+frameIdx*delta_t)
startTime = Float32(currTime)
endTime = Float32(currTime+(SEG_SIZ-1)*delta_t)
delta = Float32(delta_t)
dim = Range(startTime, endTime, delta)
node.beginSegment(startTime, endTime, dim, currSeg, -1)
# make some data for this frame
m = math.tan((2*PI*frameIdx)/NUM_FRAMES)
for i in range(WIDTH):
j = int(round((i-WIDTH/2)*m))
if j >= -HEIGHT/2 and j < HEIGHT/2:
currFrame[(j+HEIGHT/2)*WIDTH+i] = 255
# reshape the data and call putSegment
segment = Int16Array(currFrame)
segment.resize([1,HEIGHT,WIDTH])
node.putSegment(segment, -1)
The following jScope configuration allows jScope displaying the content of the sequence of frames as shown below
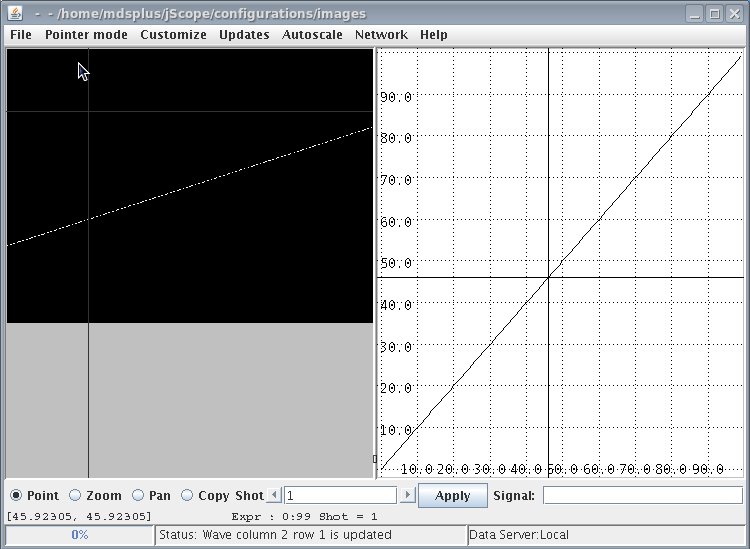
The window on the right is just a straight line representing time: when jScope is in "point" configuration, dragging the mouse along this waveform will move the frame visualization accordingly. In more realistic applications, this feature is useful to correlate the displayed images to the occurrence of interesting facts as shown by some waveform.
Scope.geometry: 750x550+272+162 Scope.update.disable: false Scope.update.disable_when_icon: true Scope.font: java.awt.Font[family=Dialog,name=Times,style=plain,size=14] Scope.color_0: Black,java.awt.Color[r=0,g=0,b=0] Scope.color_1: Blue,java.awt.Color[r=0,g=0,b=255] Scope.color_2: Cyan,java.awt.Color[r=0,g=255,b=255] Scope.color_3: Green,java.awt.Color[r=0,g=255,b=0] Scope.color_4: Magenta,java.awt.Color[r=255,g=0,b=255] Scope.color_5: Orange,java.awt.Color[r=255,g=200,b=0] Scope.color_6: Pink,java.awt.Color[r=255,g=175,b=175] Scope.color_7: Red,java.awt.Color[r=255,g=0,b=0] Scope.color_8: Yellow,java.awt.Color[r=255,g=255,b=0] Scope.data_server_name: Local Scope.data_server_class: LocalDataProvider Scope.fast_network_access: false Scope.reversed: false Scope.global_1_1.experiment: test Scope.global_1_1.horizontal_offset: 0 Scope.global_1_1.vertical_offset: 0 Scope.columns: 2 Scope.rows_in_column_1: 1 Scope.plot_1_1.height: 434 Scope.plot_1_1.grid_mode: 0 Scope.plot_1_1.is_image: true Scope.plot_1_1.keep_ratio: true Scope.plot_1_1.horizontal_flip: false Scope.plot_1_1.vertical_flip: false Scope.plot_1_1.palette: Green scale Scope.plot_1_1.bitShift: 0 Scope.plot_1_1.bitClip: false Scope.plot_1_1.num_expr: 1 Scope.plot_1_1.num_shot: 1 Scope.plot_1_1.global_defaults: 212865 Scope.plot_1_1.y_expr_1: \segmented Scope.rows_in_column_2: 1 Scope.plot_1_2.height: 434 Scope.plot_1_2.grid_mode: 0 Scope.plot_1_2.x_log: false Scope.plot_1_2.y_log: false Scope.plot_1_2.update_limits: true Scope.plot_1_2.palette: Green scale Scope.plot_1_2.bitShift: 0 Scope.plot_1_2.bitClip: false Scope.plot_1_2.num_expr: 1 Scope.plot_1_2.num_shot: 1 Scope.plot_1_2.global_defaults: 262017 Scope.plot_1_2.x_expr_1: 0:99 Scope.plot_1_2.y_expr_1: 0:99 Scope.plot_1_2.mode_1D_1_1: Line Scope.plot_1_2.mode_2D_1_1: xz(y) Scope.plot_1_2.color_1_1: 0 Scope.plot_1_2.marker_1_1: 0 Scope.plot_1_2.step_marker_1_1: 1 Scope.vpane_1: 500
You can copy this configuration in a jscp file and pass it in jScope command (or reading it in Customize->Use Saved Setting from... option).
Storing Images as Opaque objects in a node
Arbitrary strings of bytes can now be stored using an Opaque object which has two properties, the value portion consisting of a byte array and a description portion which is a string hint which can be used by applications to determine how to interpret the byte array. Opaque objects can be used to store images and movies for example. Segmented records have been extended to permit you to store a series of Opaque objects in a node. The Python module provides a getImage() method for Opaque objects which uses PIL (Python Imaging Library) to return an Image object that provides imaging methods for displaying or manipulating images. The Opaque class in Python also provides a fromFile() method which can be used to create an Opaque object from an image file. The following python example illustrates this capability:
from MDSplus import *
t=Tree('test',1,'new')
n=t.addNode('image','signal')
n.record=Opaque.fromFile('/home/twf/mygif.gif')
n2=t.addNode('imageseg','signal')
n2.makeSegment(None,None,None,Opaque.fromFile('/home/twf/mygif.gif'))
n2.makeSegment(None,None,None,Opaque.fromFile('/home/twf/mympeg.mpeg'))
n2.makeSegment(None,None,None,Opaque.fromFile('/home/twf/myjpeg.jpeg'))
t.write()
n.record.getImage().show() ### display the image in the node
n2.getSegment(0).getValue().getImage().show() ### display the image in the segment
NOTE: To use the getImage() method you need to have the PIL python module installed. To use the show() method you also need a command line tool that PIL can invoke to show the image. On linux there is an ImageMagick package than provides a display command the PIL will use.
Handling Lists of Heterogeneous Data Items
The Object Oriented Data interface allows the practical usage of a feature that has always been available in MDSplus, but which has almost never been used before, i.e. the possibility of storing structures of heterogeneous data items in tree nodes. Every component of such structure can be any MDSplus datatype, including structures themselves. A data structure gets specified via class Apd. This class represent a generic list of Data objects, including Apds (class Apd is a subclass of Data). The usage of Apd objects is shown by the following example, in which a structure composed of a String and an integer is stored in a tree node named STRUCT. In order to accept data structures, the usage of the tree node must be "ANY".
C++
Tree *tree = new Tree("my_tree", -1);
TreeNode *node = tree->getNode("STRUCT");
Data **fields = new *Data[2];
data[0] = new String("SOME TEXT HERE");
data[1] = new Int32(1);
Data *apd = new Apd(2, fields);
node->putData(apd);
Java
Tree tree = new Tree("my_tree", -1);
TreeNode node = tree.getNode("STRUCT");
Data apd = new Apd(new Data[]{new String("SOME TEXT HERE"), new Int32(1)});
node.putData(apd);
Python
n = Tree('my_tree', -1).getNode('STRUCT')
n.putData(["SOME TEXT HERE", 1])
As you can see the Python example is very concise. In fact, Python represents the best language for handling MDSplus data structures, being reflected in the Python list construct. More support for lists and dictionaries is provided in the Python interface. Refer to the section describing Python-specific issues for a more complete description.
Some working examples
In this section some complete programs written in C++, Java and python are presented, describing also the required steps for their compilation. All these programs are available in the support MDSplusTutorial project used in the first part of this tutorial. The examples are in device directory and can be built via the make command.
First example: expression evaluation
The first example provides the evaluation of a given expression. MDSplus expressions may contain only components not related to given pulse files (e.g. 2+5) or may refer to data items in pulse files (e.g. MY_DATA * 12.3). In the latter case the expression must be evaluated in the context of a given pulse file or experiment model, which must be open before evaluation the expression. This program optionally accepts the experiment name and the shot arguments to handle the latter case.
The valuation of the a MDSplus expression from its textual representation requires first its compilation (method Data.compile()) and then the evaluation of the resulting Data instance (method Data.data()). The two methods can be replaced by method Data.execute() that transforms the passed expression string into the resulting Data instance. Recall that when an expression is evaluated, the result is represented by either a scalar or array (integer, floating point or string) since all the other data types are resolved in the evaluation (for example data references in pulse files are resolved during the expression evaluation).
The c++ source code is the following:
#include <mdsobjects.h>
#include <iostream>
#include <stdlib.h>
#include <stdio.h>
/*******************************************
* Evaluation of a MDSplus expression.
* The expression is passed as first argument. An Optional second argument defines an
* experiment. a further optional argument defines the shot number.
* If only one command line argument is passed no tree is open;
* If only two command arguments are passed, shot -1 (the model) is open for the passed experiment name.
* The programs prints the decompiled form of the Data object returned by the evaluation.
********************************************/
int main(int argc, char *argv[])
{
if(argc < 2 || argc > 4)
{
std::cout << "Usage: eval_expr <expression> [experiment] [shot]" << std::endl;
exit(0);
}
char *expression = argv[1];
MDSplus::Data *evalData;
if(argc > 2)
{
char *experiment = argv[2];
int shot = -1;
if(argc > 3)
sscanf(argv[3], "%d", &shot);
MDSplus::Tree *tree;
try {
//Open the tree in a try block
tree = new MDSplus::Tree(experiment, shot);
} catch(MDSplus::MdsException &exc)
{
//methode MdsException::what() return a description of the exception
std::cout << "Cannot open tree " << experiment << " shot " << shot << ": " << exc.what() << std::endl;
exit(0);
}
try {
//MDSplus::execute first compiles the passed striing and then evaluates the expression
//It is equivalent to:
// MDSplus::Data *compiledData = MDSplus::compile(expression, tree);
// evalData = compiledData->data();
// deleteData(compiledData);
evalData = MDSplus::execute(expression, tree);
std::cout << evalData << std::endl;
} catch(MDSplus::MdsException &exc)
{
std::cout << "Cannot evaluate " << expression << ": " << exc.what() << std::endl;
exit(0);
}
// Data objects must be freed via routine deleteData()
deleteData(evalData);
// Tree object instead use C++ delete operator
delete(tree);
}
else //No experiment
{
try {
// Here the tree argument is not passed because no reference tree is open
evalData = MDSplus::execute(expression);
std::cout << evalData << std::endl;
} catch(MDSplus::MdsException &exc)
{
std::cout << "Cannot evaluate " << expression << ": " << exc.what() << std::endl;
exit(0);
}
deleteData(evalData);
}
}
Use the following commands to compile and link the source
gcc -c -I$MDSPLUS_DIR/include eval_expr.cpp g++ -o eval_expr eval_expr.o -L$MDSPLUS_DIR/lib -lMdsObjectsCppShr
The first command specifies the directopry containing the mdsobjects.h include file for C++ MDSplus API. The second one specifies the directory containing the required libraries and the library to link against. Environment variable MDSPLUS_DIR is set during the installation procedure of MDSplus.
The Java source is the following:
class eval_expr
{
public static void main(String []args)
{
if(args.length < 1 || args.length > 3)
{
System.out.println("Usage: eval_expr <expression> [experiment] [shot]");
System.exit(0);
}
String expression = args[0];
if(args.length > 1)
{
String experiment = args[1];
int shot = -1;
if(args.length > 2)
shot = Integer.parseInt(args[2]);
MDSplus.Tree tree = null;
try {
//Open the tree in a try block
tree = new MDSplus.Tree(experiment, shot);
} catch(MDSplus.MdsException exc)
{
System.out.println("Cannot open tree " + experiment +" shot " + shot + ": " + exc);
System.exit(0);
}
}
//execute first compiles the passed string and then evaluates the expression
//It is equivalent to:
// MDSplus.Data compiledData = MDSplus::compile(expression);
// evalData = compiledData.data();
MDSplus.Data evalData = MDSplus.Data.execute(expression, new MDSplus.Data[0]); //Second argument means no argument passed
if(evalData == null)
System.out.println("Cannot evaluate " + expression);
else
System.out.println(evalData);
}
}
The commands to compile and to execute the java program are the following
javac -cp $MDSPLUS_DIR/java/classes/mdsobjects.jar eval_expr.java java -cp $MDSPLUS_DIR/java/classes/mdsobjects.jar:. eval_expr <expression> [experiment name] [shot]
Java requires the definition of the CLASSPATH environment variable to include the class definition of MDSplus (in mdsobjects.jar). When launched, it is also necessary to include the current directory so that the compiled eval_expr class can be found by the Java Virtual Machine.
Finally, the python source is shown below
import sys
from MDSplus import *
if(len(sys.argv) < 2 or len(sys.argv) > 4):
print "Usage: eval_expr <expression> [experiment] [shot]";
sys.exit(0);
if(len(sys.argv) > 2):
experiment = sys.argv[2];
shot = -1;
if(len(sys.argv) > 3):
shot = int(sys.argv[3]);
try:
tree = Tree(experiment, shot)
except:
print "Cannot open tree " + experiment +" shot " + shot
sys.exit(0);
expression = sys.argv[1]
try:
evalData = Data.execute(expression);
print evalData
except:
print "Cannot evaluate expression "+ expression
The python program is invoked by the command
python eval_expr.py
The three examples above show how, apart from unavoidable differences due to the the different programming environments (such as the need of explicitly deallocating objects in C++), the used classes are quite similar across languages.
The presented example is extended a bit in the next one to show how it is possible to inspect data classes.
Second example: expression evaluation and resulting data inspection
In this second example, the resulting data instance obtained by the evaluation of the expression is inspected in order to detect whether the result is a scalar or an array, and the content displayed, without using the default decompilation of the expression in print command. As the first part of the example is the same of the first one, only the changed part (after execute() method invocation) is displayed below in the three considered languages. The complete source files are available in the MDSplusTutorial project.
C++
....
//evalData is the result of the expression evaluation. In order to render it, inquire it via getInfo method
//method getInfo returns information about the data object in a C like approach.
char dataClass, dataType, nDims; //Returned data type, class and number of dimensions
short dataSize; //Returned total data size in bytes
int *dims; //Returned array dimensions
void *dataPtr; //Returned internal data pointe. NEVER use it!!
evalData->getInfo(&dataClass, &dataType, &dataSize, &nDims, &dims, &dataPtr);
//Returned dataClass for evalated data will be either CLASS_S (scalar) or CLASS_A (array)
int nItems = 1;
switch(dataClass) {
case CLASS_S: //Scalar
if(dataType == DTYPE_FLOAT || dataType == DTYPE_DOUBLE) //If the returned scalar is floating point
std::cout << "Scalar: " << evalData->getDouble() << std::endl;
else if(dataType == DTYPE_T) //If the returned type is a string
{
char *retStr = evalData->getString();
std::cout << "Scalar: " << retStr << std::endl;
delete [] retStr; //Deallocate returned string
}
else //The returned data is integer
std::cout << "Scalar: " << evalData->getInt() << std::endl;
break;
case CLASS_A: //array
std::cout << "Array ( ";
for(int i = 0; i < nDims; i++)
{
nItems *= dims[i];
std::cout << dims[i] << " ";
}
std::cout << ") " << std::endl;
if(dataType == DTYPE_FLOAT || dataType == DTYPE_DOUBLE)
{
double *retDoubleArr = evalData->getDoubleArray(&nItems);
for(int i = 0; i < nItems; i++)
std::cout << retDoubleArr[i] << " ";
delete [] retDoubleArr; //Deallocate returned native array
}
else if (dataType == DTYPE_T) //String Array
{
char **strings = evalData->getStringArray(&nItems);
for(int i = 0; i < nItems; i++)
{
std::cout << "\"" << strings[i] << "\" ";
delete [] strings[i];
}
delete [] strings;
}
else //Integer array
{
int *retIntArr = evalData->getIntArray(&nItems);
for(int i = 0; i < nItems; i++)
std::cout << retIntArr[i] << " ";
delete [] retIntArr; //Deallocate returned native array
}
break;
default:
std::cout << "Unexpected data class" << std::endl;
}
deleteData(evalData); //Deallocate evaluated data object
return 0;
}
Java
....
//evalData is the result of the expression evaluation.
if(evalData instanceof MDSplus.Scalar)
System.out.println("Scalar: "+evalData); //Use the default toString() method for scalar data
else if(evalData instanceof MDSplus.Array)
{
try {
int dims[] = evalData.getShape();
System.out.print("Array ( ");
for(int i = 0; i < dims.length; i++)
System.out.print(dims[i] + " " );
System.out.println(")");
if(evalData instanceof MDSplus.Float32Array || evalData instanceof MDSplus.Float64Array)
{
float[] data = evalData.getFloatArray();
for(int i = 0; i < data.length; i++)
System.out.print(data[i] + " " );
}
else if(evalData instanceof MDSplus.StringArray)
{
String[] data = evalData.getStringArray();
for(int i = 0; i < data.length; i++)
System.out.print("\""+data[i] + "\" " );
}
else //Integer
{
int[] data = evalData.getIntArray();
for(int i = 0; i < data.length; i++)
System.out.print(data[i] + " " );
}
}catch(MDSplus.MdsException exc) { System.out.println("Cannot display evaluated data: " + exc);}
}
else
System.out.println("Unexpected returned data type");
}
}
python
...
if(isinstance(evalData, Scalar)):
print "Scalar: " + evalData
else:
if(isinstance(evalData, Array)):
print "Array", evalData.getShape()
if (isinstance(evalData, Float32Array) or isinstance(evalData, Float64Array)):
print evalData.getFloatArray()
else:
if (isinstance(evalData, StringArray)):
print evalData.getString()
else:
print evalData.getIntArray()
Observe that, unlike java and python, C++ is lacking an introspection mechanism for classes, and therefore a C++ specific method getInfo() is provided in order to query the current data instance, getting information about the actual data type and whether data is a scalar (i.e. subclass of Scalar) or array (i.e. subclass of Array).
Third example: traversing the tree structure of a pulse file
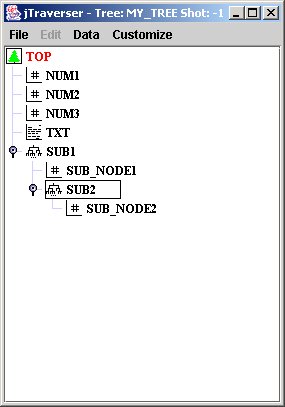
In this example an experiment name and an optional shot are passed to the program which traverses the hierarchical structure of the pulse file and prints the names of the traversed nodes. You can try it with the tutorial pulse file available in the tutorial project, but recall first that the environment variable tutorial_path must be defined to the name of the directory containing the pulse files (<MDSplusTutorial home/trees>).
The C++ source is the following:
#include <mdsobjects.h>
#include <iostream>
#include <stdlib.h>
/*************************************************
* Dump the tree structure of a MDSplus pulse file. The program will print
* the name of every node in the tree aligned to highlight the tree hierarchy
*
*************************************************/
//Recursive traversal routine
static void traverseTree(MDSplus::TreeNode *rootNode, int tabs);
int main(int argc, char *argv[])
{
if(argc < 2 || argc > 4)
{
std::cout << "Usage: dump_tree <experiment> [shot]" << std::endl;
exit(0);
}
char *experiment = argv[1];
int shot = -1;
if(argc > 2)
sscanf(argv[2], "%d", &shot); //If no shot specified, the experiment model (shot -1) is considered
MDSplus::Tree *tree;
try {
tree = new MDSplus::Tree(experiment, shot);
} catch(MDSplus::MdsException &exc)
{
std::cout << "Error opening tree " << experiment << ": " << exc.what() << std::endl;
exit(0);
}
//Get the top node. It is named by default "\TOP"
MDSplus::TreeNode *topNode = tree->getNode("\\TOP");
traverseTree(topNode, 0);
//Deallocate topNode and tree
delete topNode;
delete tree;
return 0;
}
//Recursive tree traversal
static void traverseTree(MDSplus::TreeNode *rootNode, int tabs)
{
//Print the root name, get the list of the descendats and recurse on them
//getNodeName() returns a C string which must be dealloaed afterwards
char *rootName = rootNode->getNodeName();
for(int i = 0; i < tabs; i++)
std::cout << "\t";
std::cout << rootName << std::endl;
delete []rootName;
// Get all the descendant (members + children)for this node
int numDescendants;
MDSplus::TreeNode **descendants = rootNode->getDescendants(&numDescendants);
//Traverse hierarchy
for(int i = 0; i < numDescendants; i++)
traverseTree(descendants[i], tabs + 1);
//Deallocate the returned array of tree nodes
for(int i = 0; i < numDescendants; i++)
delete descendants[i];
delete [] descendants;
}
The program is compiled and linked using the following commands:
gcc -c -I$MDSPLUS_DIR/include dump_tree.cpp g++ -o dump_tree -L$MDSPLUS_DIR/lib dump_tree.o -lMdsObjectsCppShr
The Java code is the following:
class dump_tree
{
public static void main(String args[])
{
if(args.length < 1 || args.length > 3)
{
System.out.println("Usage: dump_tree <experiment> [shot]");
System.exit(0);
}
String experiment = args[0];
int shot = -1;
if(args.length > 1)
shot = Integer.parseInt(args[1]);
MDSplus.Tree tree = null;
try {
tree = new MDSplus.Tree(experiment, shot);
} catch(MDSplus.MdsException exc)
{
System.out.println("Error opening tree " + experiment + ": " + exc);
System.exit(0);
}
//Get the top node. It is named by default "\TOP"
try {
MDSplus.TreeNode topNode = tree.getNode("\\TOP");
traverseTree(topNode, 0);
}catch(MDSplus.MdsException exc)
{
System.out.println("annot traverse tree: " + exc);
}
}
//Recursive tree traversal
static void traverseTree(MDSplus.TreeNode rootNode, int tabs) throws MDSplus.MdsException
{
//Print the root name, get the list of the descendats and recurse on them
//getNodeName() returns a C string which must be dealloaed afterwards
String rootName = rootNode.getNodeName();
for(int i = 0; i < tabs; i++)
System.out.print("\t");
System.out.println(rootName);
MDSplus.TreeNodeArray descendants = rootNode.getDescendants();
//Traverse hierarchy
for(int i = 0; i < descendants.size(); i++)
traverseTree(descendants.getElementAt(i), tabs + 1);
}
}
The Java program is compiled and executed using the following commands:
javac -cp $MDSPLUS_ROOT/java/classes/mdsobjects.jar dump_tree,java java -cp $MDSPLUS_ROOT/java/classes/mdsobjects.jar:. dump_tree <experiment name>[shot]
Finally, the python code is the following:
import sys
from MDSplus import *
def traverseTree(rootNode, tabs):
rootName = rootNode.getNodeName()
for i in range(0, tabs):
print "\t",
print rootName
try:
children = rootNode.getChildren()
for c in children:
traverseTree(c, tabs+1)
except:
pass
try:
members = rootNode.getMembers()
for m in members:
traverseTree(m, tabs+1)
except:
pass
if(len(sys.argv) < 2 or len(sys.argv) > 3):
print "Usage: dump_tree <experiment> [shot]"
sys.exit(0)
experiment = sys.argv[1]
shot = -1
if(len(sys.argv) > 2):
shot = int(sys.argv[2])
try:
tree = Tree(experiment, shot)
except:
print "Cannot open tree " + experiment +" shot " + shot
sys.exit(0);
topNode = tree.getNode("\\TOP")
traverseTree(topNode, 0);
For the braves: deep copy of pulse files
In MDSplusTutorial project there is a fourth example: copy_tree.cpp which performs a deep copy of a given pulse file. That program traverses the structure of the input pulse file and produces a new pulse file, building it step by step which the first pulse file is traversed. This program takes into account more specialized aspects of MDSplus and therefore you can easily skip this section in case you are going to use MDSplus just for reading data in pulse files.
In the first step the program recursively traverse the input pulse file, building the output pulse file structure. When this step is finished, the output pulse file, open in edit mode, is closed and re-open in normal mode because it is not necessary to open a pulse file in edit mode in order to read and write data items.
In the second step, the input and output pulse files are recursively traversed and data items from the input pulse file are copied in the second one. There are however several facts which must be taken into account when creating and populating the output pulse file:
- Management of subtrees pulse file can contain reference to other linked pulse files, which are automatically opened and traversed when accessing the links. This [program however copy only the given tree, and therefore recursion must be stopped when a node which represents the root of an externally linked subtree is found. This is performed by inquiring the Usage of that node.
- Creation of Device subtrees MDSplus devices are explained in Developing MDSplus Devices section and require that the subtree corresponding to a device instance is not created node by node in the usual way, rather delegating the creation of the corresponding subtree to the device constructor method.
- Copy of segmented nodes we have seen that data items may containing normal data, represented by expressions, or segmented data, that is, a sequence of segment, each holding a part of the a signal. The program must therefore detect whether the data item is a segmented one and in this case perform the copy segment by segment.
- Management of tree node references in expressions we have seen that expression may contain references to data items in pulse files. Such references are either represented by a TreeNode or a TreePath object instances in the corresponding data hierarchy. A TreeNode instance maintains the reference in the for of an internal identifier (an integer number) which is however valid only in the context of the target tree, and may be different in another tree. A TreePath reference maintains the reference under the form of a path name, and is therefore valid in the context of any tree. When copying expressions from one tree to another, it is necessary to make sure that all TreeNode instances are replaced by equivalent TreePath ones, by traversing the hierarchy of Data objects representing the expression and making the required replacements.
You can exercise the program in MDSplus tutorial project using the tutorial tree giving the usual commands for compilation and link:
gcc -c -I$MDSPLUS_DIR/include copy_tree.cpp g++ -o copy_tree -L$MDSPLUS_DIR/lib copy_tree.o -lMdsObjectsCppShr
however, before running the program, e.g. with the command
copy_tree tutorial 1 tutorial 10
which copies shot 1 into shot 10, you need to create the necessary infrastructure for the management of the device (DEMO_ADC) declared in that pulse file. Without entering into details (interested readers may look at Developing MDSplus Devices section) it suffices giving (once for all) the following commands for the installation of the DEMOADC device used in the tutorial:
cd $MDSPLUS_ROOT/tdi/TutorialDevices python setup.py install
Java Specific Issues
The MDSplus classes for Java are contained in the MDSplus package. Therefore it is necessary to import the package classes via the following line:
import MDSplus.*;
There is a potential name clash for the String class. When referring to Java Strings, it is therefore necessary to provide the fully specified definition, i.e. java.lang.String, while for MDSplus strings, the class name is MDSplus.String. String values returned by TreeNode (e.g. method getPath())always refer to java.lang.String.
Error management is achieved using exceptions. An instance of class MdsException is thrown when an error occurs. It is therefore necessary to include Tree and TreeNode methods within a try block, as shown in the following example:
try {
t = Tree("my_tree", -1);
TreeNode n = tree.getNode('data');
}catch(MdsException exc)
{
System.out.println("MDSplus error: " + exc);
}
A complete reference to the Java classes and methods for MDSplus is available here.
C++ Specific Issues
File mdsobjects.h contains all the definitions of the C++ MDSplus classes.
The C++ classes for MDSplus objects are declared to belong to MDSplus namespace. It is therefore convenient to begin the program as follows:
#include <mdsobjects.h> using namespace MDSplus;
It is possible to avoid the second statement above and refer always to the MDSplus namespace, for example:
MDSplus::Tree = new MDSplus::Tree("my_tree", -1);
When native arrays are passed to Data constructors (a.g. when instantiating Array classes) , the passed array is copied in class-private storage. When native arrays are returned by Data objects (e.g. in the getXXXArray() methods), they are allocated by the class methods using the new operator, and copied from internal object fields. Returned arrays need therefore to be deallocated by the calling program via delete [] operator when they are no longer needed. Note however that on Windows deleting an array which has been allocated in a different DLL may crash the program. For this purpose, use always routine deleteNativeArray() to free arrays returned by accessor methods. When a method returns a native array, its pointer is directly returned by the method, while the number of elements is returned via an argument, passed by reference.
In order to ease the management of dynamic memory for object allocation and deallocation, and to reduce the risk of memory errors and leaks, the following approach is recommended:
- Allocate always MDSplus objects using operator new. Do not declare MDSplus objects as local variables, but work only with pointers
- Do not use the free operator directly, use function deleteData() instead. When using deleteData() function for deallocating MDSplus objects, it is required to deallocate only those objects which have not been passed to any other constructor. Consider the following code example:
Data *start = new Float64(2.); Data *range = new RangeData(start, new Float64(3.), new Float64(1E-3)); ... deleteData(range),
assuming that range has not been passed to any other constructor, only variable range has to be deallocated. Function deleteData() handles internally reference counts for linked Data instances, so that objects are deallocated when no longer needed. For this reason no memory leaks occur in the example above.
Data objects returned by getter methods of other data objects must be deallocated via function deleteData() when no longer needed.
In summary, you are required to call deleteData() for a Data object no longer needed when either condition hold:
- The object has been allocated via the new operator and has not been passed to any other Data constructor or setter method of Data classes;
- The object has been obtained by a getter method of Data classes.
Non Data classes (such as Tree, TreeNode etc) are instead freed via the usual delete operator.
Arrays returned by every accessor method have to be deallocated using routine deleteNativeArray().
Error management in C++ is handled using exceptions. An instance of MdsException is thrown when an error occurs, bringing the description of the error. It is therefore necessary to define a try block when using Tree and TreeNode methods, as shown in the following example:
try {
Tree *tree = new Tree("my_tree", -1);
TreeNode *node = tree->getNode("data");
} catch(MdsException *exc)
{
cout << "MDSplus error: " << exc << "\n";
}
Python Specific Issues
Local use of MDSplus objects in python
To use the MDSplus objects in Python you must import them using a command such as:
from MDSplus import *
The Python implementation, unlike the other languages, allows you to reference tree node characteristics as class instance properties. If you are familiar with the MDSplus TDI function GETNCI, then any of the items you might request using GETNCI can be referenced as a property.
mynode = Tree("mytree", -1).getNode("mynode")
length = mynode.length
data = mynode.record
dtype = mynode.dtype
name = mynode.node_name
path = mynode.path
on = mynode.on
The methods for accessing these properties are also available for compatibility with other language implementations:
length = mynode.getLength() data = mynode.getData() dtype = mynode.getDtype() name = mynode.getNodeName() path = mynode.getPath() on = mynode.isOn()
Another shortcut found in the Python implementation is the ability to store data in a node using the record property:
mynode.record = 42
It is handy to operate on MDSplus datatypes directly, i.e. without calling the data() method to extract a Python-native object, as mentioned above. In certain situations, however, it is necessary to be aware of the difference. For example, the ** operator is not implemented for (numeric) MDSplus datatypes. Another (non-trivial) difference is array slicing:
a = Float64Array([2.72, 3.14, 6.02]) b = a.data() a[0:1] ### TDI/IDL convention [2.72D0,3.14D0] b[0:1] ### Python convention array([ 2.72])
MDSplus also allows you to store python dictionary objects into MDSplus trees as long as the values in the dictionary object are of a type supported by MDSplus. The following are some examples of storing dictionaries:
mynode.record={'f1':32,'f2':42} ### a simple dictionary with two fields each with int values.
mynode.record={'field1':42,'myfield2':'a string','mydict':{'f1':42,'f2':43.}} # nested dicts
These dictionary objects can be referenced using TDI as well:
TDI> MYNODE['mydict']['f1'] 42
Remote interface to MDSplus objects using ssh
You can now use the MDSplus object interface connecting to a remote system using SSH. You do not need any of the MDSplus software packages installed on your local computer to use this feature. There are a couple caveats with this approach which will be mentioned below but nearly all the functionality of the MDSplus objects can be obtained using a small python module called mdsconnector available using pip install.
To install this package you need to use the pip python utility. Either of the two commands should work if pip is installed on your local computer:
$ pip install mdsconnector $ python -m pip install mdsconnector
Once installed you can import the module and make a connection to a remote host:
$ python from mdsconnector import mdsConnector c = mdsConnector(host-name[,user=username,password=password])
If no username or password is provided the ssh connection will use the local account name and try ssh keys and if necessary prompt for a password.
Once you have the connection object you can reference MDSplus objects using that connection object. For example:
tree=c.Tree('tree-name',shot)
node=c.getNode('node-spec')
data=node.record.data()
Caveats
Windows
Currently this does not work well on a Windows system. This connection object uses the rpyc and plumbum python modules and on windows it attempts to use Putty ssh tools. The newer version of Windows 10 now includes OpenSSH ssh and scp but the plumbum module does not currently support this.
Output from functions such as tcl()
Functions and methods that normally write to stdout or stderr do not seem to produce output on the client machine. Many of these functions provide options to specify variables to set to capture the output text so you could then print it locally on the client.
Access privileges
Your connection will obtain the access privileges on the remote system based on the account you are ssh'ing to, essentially the same as if you manually used ssh to connect to the remote system and entered python on that system.
Environment
The environment variables on the remote system will be used for things like creating Tree instances. If your login environment on the remote system does not automatically define various tree path environment variables you will have to define them manually using the setenv function of the connection object:
c=mdsConnector('myremotehost')
c.setenv('mytree_path','/trees/mytree')
Advanced Options
It is possible to specify a hop in the mdsConnection() arguments which will cause the connection to first connect to the specified remote system and then from that system to connect to another system accessible from the first system. Some facilities use a gateway host for remote connections but the data access is provided on other local systems accessible only from the gateway host. You can find some information on the optional arguments for the mdsConnector object by doing the following:
from mdsconnector import mdsConnector help(mdsConnector)
MATLAB Specific Issues
MATLAB can make a straightforward use of all the presented classes using their Java interface. Installation is a bit tricky, the names and locations of the configuration files vary with MATLAB version and platform. In addition the exact location of the MDSplus files that need to be in the configuration also varies by platform, and in some cases release.
Configuration
- add the MDSplus directory containing the MATLAB '.m' files to the the MATLAB path.
- make sure the env MDSPLUS_DIR is set to the MDSplus installation path, e.g. "/usr/local/mdsplus" (linux) or "%ProgramFiles%\MDSplus" (Windows)
MATLAB>> addpath(fullfile(getenv('MDSPLUS_DIR'),'matlab')); savepath; % adds the path to current list and makes current list default
On modern version of Matlab you are now ready to go. Older version require a few more steps.
Older versions of Matlab
- add the path to the "mdsobjects.jar" file to the "javaclasspath.txt" file.
MATLAB>> system(sprintf('echo %s>>%s',fullfile(getenv('MDSPLUS_DIR'),'java','classes','mdsobjects.jar'),fullfile(prefdir,'javaclasspath.txt')));
- add the location of the mdsplus shared images for your platform to the javalibrarypath.txt file.
- linux: /usr/local/mdsplus/lib or /usr/local/mdsplus/lib64 or /usr/local/mdsplus/lib32 based on your distribution and MATLAB version.
MATLAB>> system(sprintf('echo %s>>%s','/usr/local/mdsplus/lib64',fullfile(prefdir,'javalibrarypath.txt')));
- Windows (AllUser): C:\Windows\system32 for 64-bit Matlab or C:\Windows\syswow64 for 32-bit MATLAB.
MATLAB>> system(sprintf('echo %s>>%s',fullfile(getenv('SystemRoot'),'System32'),fullfile(prefdir,'javalibrarypath.txt')));
- Windows (User): %MDSPLUS_DIR%\bin64 for 64-bit Matlab or %MDSPLUS_DIR%\bin32 for 32-bit MATLAB.
MATLAB>> system(sprintf('echo %s>>%s',fullfile(getenv('MDSPLUS_DIR'),'bin64'),fullfile(prefdir,'javalibrarypath.txt')));
- for older version of matlab need to edit the file toolbox/local/classpath.txt inside your MATLAB installation folder instead, e.g. C:\Program Files (x86)\MATLAB\R2012a\toolbox\local\classpath.txt
! You have to restart MATLAB for these changes to take effect !
Examples
Object Interface
- add the MDSplus java classes to MATLAB
>> import MDSplus.*
- create a tree object - the tree test must already be defined. For these examples it has one child node called 'foo', which is usage 'numeric'.
>> t = Tree('test', -1)
t =
Tree(test, -1, NORMAL)
- get the node object for the child called 'foo'
>> n = t.getNode('foo')
n =
FOO
- get that object's data.
>> a = n.getData() a = [[1D0], [2D0], [3D0], [4D0], [5D0], [6D0], [7D0]]
- Inspect it
>> class(a) ans = MDSplus.Float64Array
- convert it to a native MATLAB type
>> b = mdsToMatlab(a)
b =
1 2 3 4 5 6 7
>> class(b)
ans =
double
>> size(b)
ans =
1 7
- write a native array of doubles into the node, read it back and inspect it.
>> n.putData([1,2,3,4,5,6])
>> a = n.getData()
a =
[1D0,2D0,3D0,4D0,5D0,6D0]
>> class(a)
ans =
MDSplus.Float64Array
>> b = mdsToMatlab(a)
b =
1 2 3 4 5 6
>> class(b)
ans =
double
* the funny original data was put in by converting the native 1d array to an MDS native data type before writing. This is optional for 1d data, but required for multi dimensional arrays.
>> n.putData(mdsFromMatlab(b))
>> a = n.getData()
a =
[[1D0], [2D0], [3D0], [4D0], [5D0], [6D0]]
>> b = mdsToMatlab(a)
b =
1 2 3 4 5 6
>> class(b) ans = double
- write a two dimensional array into the node, read it back and inspect it.
>> m = magic(4)
m =
16 2 3 13
5 11 10 8
9 7 6 12
4 14 15 1
>> n.putData(mdsFromMatlab(m))
>> a = n.getData()
a =
[[16D0,5D0,9D0,4D0], [2D0,11D0,7D0,14D0], [3D0,10D0,6D0,15D0],[13D0,8D0,12D0,1D0]]
>> class(a)
ans =
MDSplus.Float64Array
>> b = mdsToMatlab(a)
b =
16 2 3 13
5 11 10 8
9 7 6 12
4 14 15 1
>> class(b)
ans =
double
Procedural Interface
An equivalent procedural interface to the one previously provided via mex, and for the IDL language is also available layered on top of these objects. The routines that make up this API are:
- [mdsconnect('host')]
- mdsopen('tree-name', shot-number)
- mdsvalue('expression' [, arguments to substitute into expression])
- mdsput('node-name', 'expression' [, arguments to substitute into expression])
- mdsclose()
- [mdsdisconnect()]
mdsconnect - mdsdisconnect provide access to thin client mdsplus hosted by a server
Examples:
Open a tree. On success return the shot number. Shot -1 is the model shot.
>> mdsopen('test', -1)
ans =
-1
Read the value from the node foo. This will return a MATLAB data structure that mimics the data stored in the node of the tree.
>> val = mdsvalue('foo');
>> whos('val')
Name Size Bytes Class Attributes
val 64x1 512 double
Write a 2x3 array into the node foo. Note the expression is simply '$' and the MATLAB array is substituted for it and written into the node. Note that odd answers (low bit set) are success.
>> x=rand(2,3);
>> whos x
Name Size Bytes Class Attributes
x 2x3 48 double
>> mdsopen('test', -1)
ans =
-1
>> mdsput('foo', '$', x)
ans =
265388041
Read back the data just written. >> mdsput('foo', '$', x)
ans =
265388041
>> val = mdsvalue('foo');
>> whos val
Name Size Bytes Class Attributes
val 2x3 48 double
Row vs Column major representation
As can be seen from the previous example array ordering is preserved across reads and writes. If a 2x3 array is written to the tree, if the node is read back a 2x3 array is returned. However, MATLAB naturally uses column-major/matrix ordering while MDSplus subscripts in row-major order. In addition, MATLAB array indices start at 1 while MDSplus array indices start at 0. For example:
>> x=rand(2,3)
x =
0.6892 0.4505 0.2290
0.7482 0.0838 0.9133
>> x(:)
ans =
0.6892
0.7482
0.4505
0.0838
0.2290
0.9133
This is preserved when this array is written to a tree and read back
>> mdsopen('test', -1)
ans =
-1
>> mdsput('foo', '$', x)
ans =
265388041
>> val=mdsvalue('foo')
val =
0.6892 0.4505 0.2290
0.7482 0.0838 0.9133
Subscripting in MATLAB the first index is the row number, and the second index is the column number
>> val(1,:)
ans =
0.6892 0.4505 0.2290
>> val(:,1)
ans =
0.6892
0.7482
However MDSplus treats this array as row major indexed starting at zero:
>> mdsvalue('foo[*,0]')
ans =
0.6892
0.7482
>> mdsvalue('foo[0,*]')
ans =
0.6892 0.4505 0.2290