From MdsWiki
|
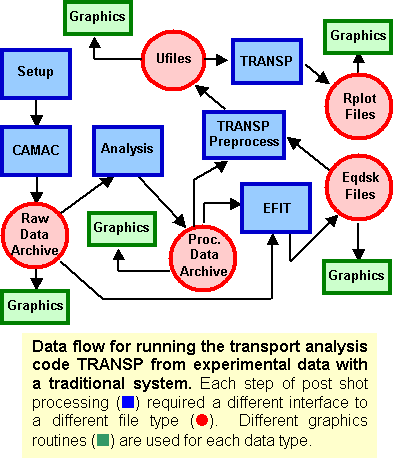
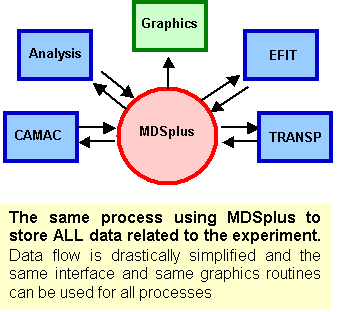
MDSplus was designed to give researchers the ability to produce complete, coherent, self-descriptive data sets and to provide tools for efficient access to that data in a distributed computing environment. The result was a system with sufficient flexibility and extensibility that ALL information associated with an experiment (or simulation) can be stored in the same structure and accessed through the same set of simple calls. By unifying setup, calibration, geometry and so forth with task descriptions, scheduling, status and all raw and processed data, MDSplus makes it easy to share data and tools across applications, facilitating collaborative research and reducing duplication of efforts. Data Driven Applications With MDSplus, it becomes practical to remove data entirely from codes and move them into the data structures. This approach allows highly flexible applications to be created whose operation can be modified easily without re-writing code. For example, a routine can create a menu of signals for display or processing by interacting with the data. New signals appear automatically in the application when they are added to the data structure - no new programming is required. Similarly, the names of routines for analysis and the parameters to control their operation can be specified as MDSplus data. Steps used during interactive processing can be saved for future reference or used as a script to "replay" the analysis. Perhaps the most important data driven application supported by MDSplus is data acquisition. Composite structures called DEVICES are defined which contain all the setup parameters, task descriptions, scheduling information and raw data which are associated with each physical data acquisition module on an experiment. Tools for scheduling and dispatching the data acquisition and analysis tasks are also part of MDSplus. A New Paradigm For Data Analysis MDSplus enables a new paradigm for post-shot processing of experimental (or simulation) data. Traditionally each processing step defined its own set of file structures, interfaces and graphical routines. For example, figure A is a simplified diagram of the data flow for a particular type of analysis (TRANSP) carried out on experimental data in the fusion program using older methodologies. The data flow resembles a multi-threaded conveyor belt spawning an array of mismatched data structures. The proliferation of file types and interface standards (APIs) make it difficult to share tools or to directly compare the results of different analysis suites. Using MDSplus, data at all stages is stored in the same format with the same API and can be accessed and displayed with the same set of routines. Data from all sources can be easily compared and tools are readily shared. Comparison With Other Data Standards MDSplus is not simply a data standard as it provides support for data acquisition, task description, scheduling and dispatching, asynchronous event generation and distribution, etc. However, MDSplus is a highly capable data storage and management system with a unique combination of features and functions including:
|